J.M. Hayes, L.C. Anderson, J.A. Schultz, M.V. Ugarov, T.F. Egan, E.K. Lewis, V. Womack, A. S. Woods, S. N. Jackson, R. H. Hauge, K. Kittrell, S. Ripley, K. K. Murray, “Matrix Assisted Laser Desorption Ionization Ion Mobility Time-of-Flight Mass Spectrometry of Bacteria,” in: C. Fenselau, P. Demirev (Eds.), Rapid Characterization of Microorganisms by Mass Spectrometry, American Chemical Society, Washington, DC, 2011: pp. 143–160. doi:10.1021/bk-2011-1065.ch009.
Abstract

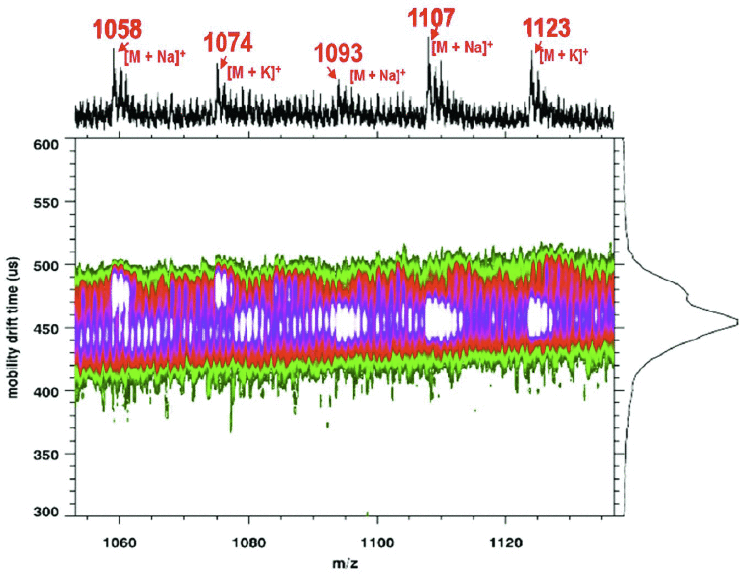
Ion mobility mass spectrometry (IMMS) was combined with matrix assisted laser desorption ionization (MALDI) for the analysis of whole cell bacteria. Whole cell Bacillus subtilis ATCC 6633 and Escherichia coli strain W ATCC 9637 bacteria were prepared with a 1:2 analyte to matrix (CHCA) ratio and deposited using the dried-droplet method. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) studies and matrix assisted laser desorption ionization ion mobility time-of-flight mass spectrometry (MALDI-IM-TOF MS) were conducted in parallel to assess the effectiveness of MALDI-IM-TOF MS for microorganism identification. Ribosomal proteins from Escherichia coli strain W ATCC 9637 were observed and assigned using the Rapid Microorganism Identification Database. Isoforms of lipopeptide products and a lantibiotic from the Bacillus subtilis ATCC 6633 species were found in the range 1000–3500 m/z and were identified using MALDI-IM-TOF MS and compared to MALDI-TOF MS data for confirmation. Vacuum ultraviolet (VUV) post-ionization MALDI-IM-TOF MS showed that additional information on lipopeptides could be obtained and used for identification.