N. Mehta, S. Shaik, A. Prasad, A. Chaichi, S.P. Sahu, Q. Liu, S.M.A. Hasan, E. Sheikh, F. Donnarumma, K.K. Murray, X. Fu, R. Devireddy, M.R. Gartia, Multimodal Label‐Free Monitoring of Adipogenic Stem Cell Differentiation Using Endogenous Optical Biomarkers, Adv. Funct. Mater., (2021) 2103955; doi: 10.1002/adfm.202103955
Abstract
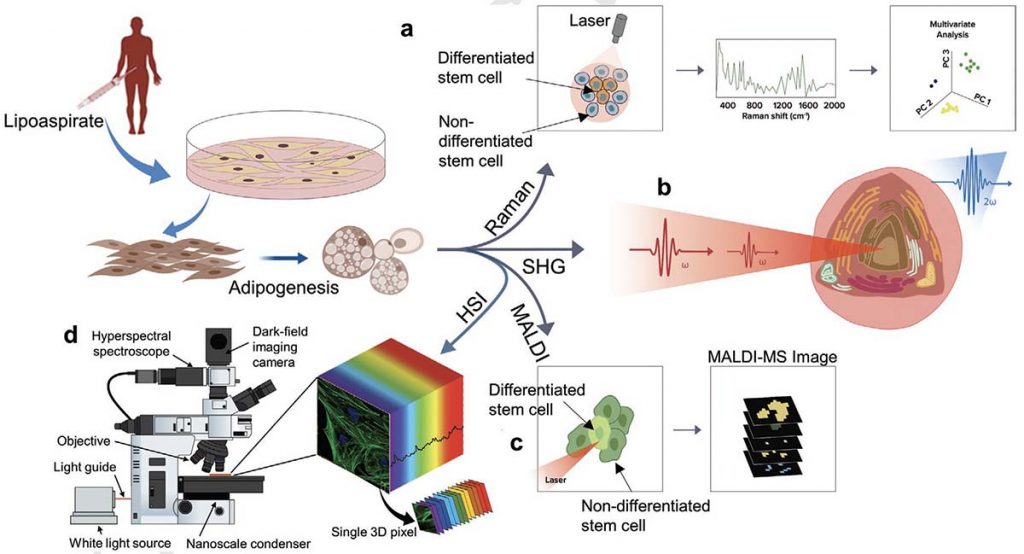
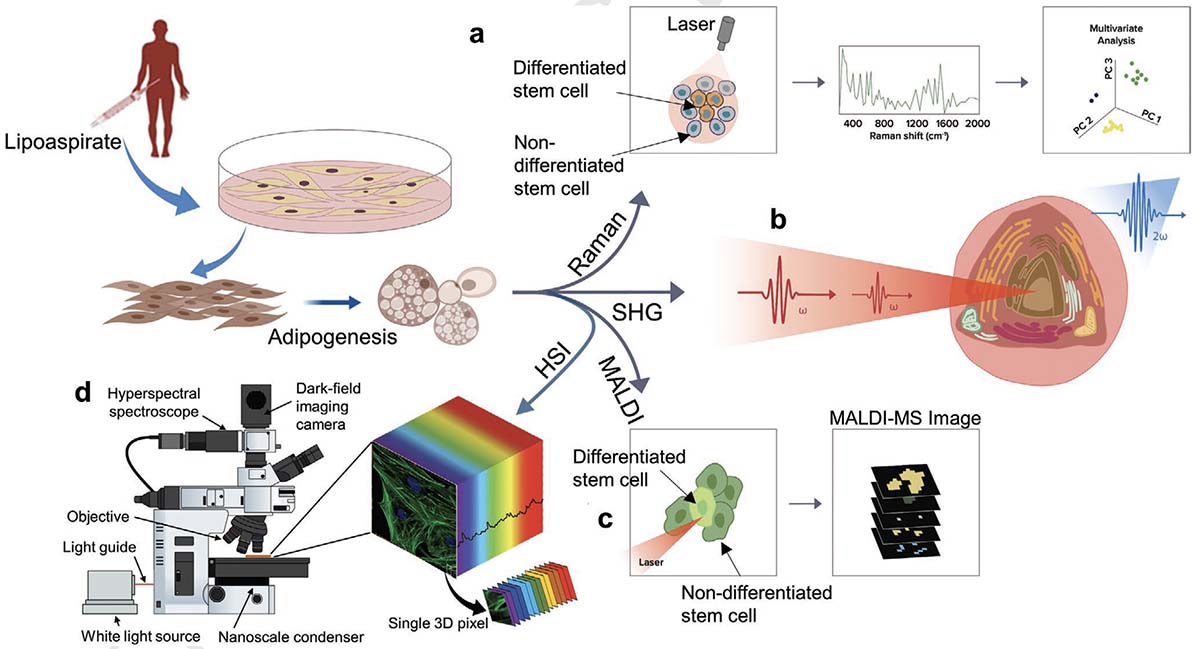
Stem cell-based therapies carry significant promise for treating human diseases. However, clinical translation of stem cell transplants for effective treatment requires precise non-destructive evaluation of the purity of stem cells with high sensitivity (<0.001% of the number of cells). Here, a novel methodology using hyperspectral imaging (HSI) combined with spectral angle mapping-based machine learning analysis is reported to distinguish differentiating human adipose-derived stem cells (hASCs) from control stem cells. The spectral signature of adipogenesis generated by the HSI method enables identifying differentiated cells at single-cell resolution. The label-free HSI method is compared with the standard techniques such as Oil Red O staining, fluorescence microscopy, and qPCR that are routinely used to evaluate adipogenic differentiation of hASCs. HSI is successfully used to assess the abundance of adipocytes derived from transplanted cells in a transgenic mice model. Further, Raman microscopy and multiphoton-based metabolic imaging is performed to provide complementary information for the functional imaging of the hASCs. Finally, the HSI method is validated using matrix-assisted laser desorption/ionization-mass spectrometry imaging of the stem cells. The study presented here demonstrates that multimodal imaging methods enable label-free identification of stem cell differentiation with high spatial and chemical resolution.