R.O. Lawal, L.T. Richardson, C. Dong, F. Donnarumma, T. Solouki, K.K. Murray, Deep-ultraviolet laser ablation sampling for proteomic analysis of tissue, Anal. Chim. Acta, 1184 (2021). doi: 10.1016/j.aca.2021.339021
Abstract
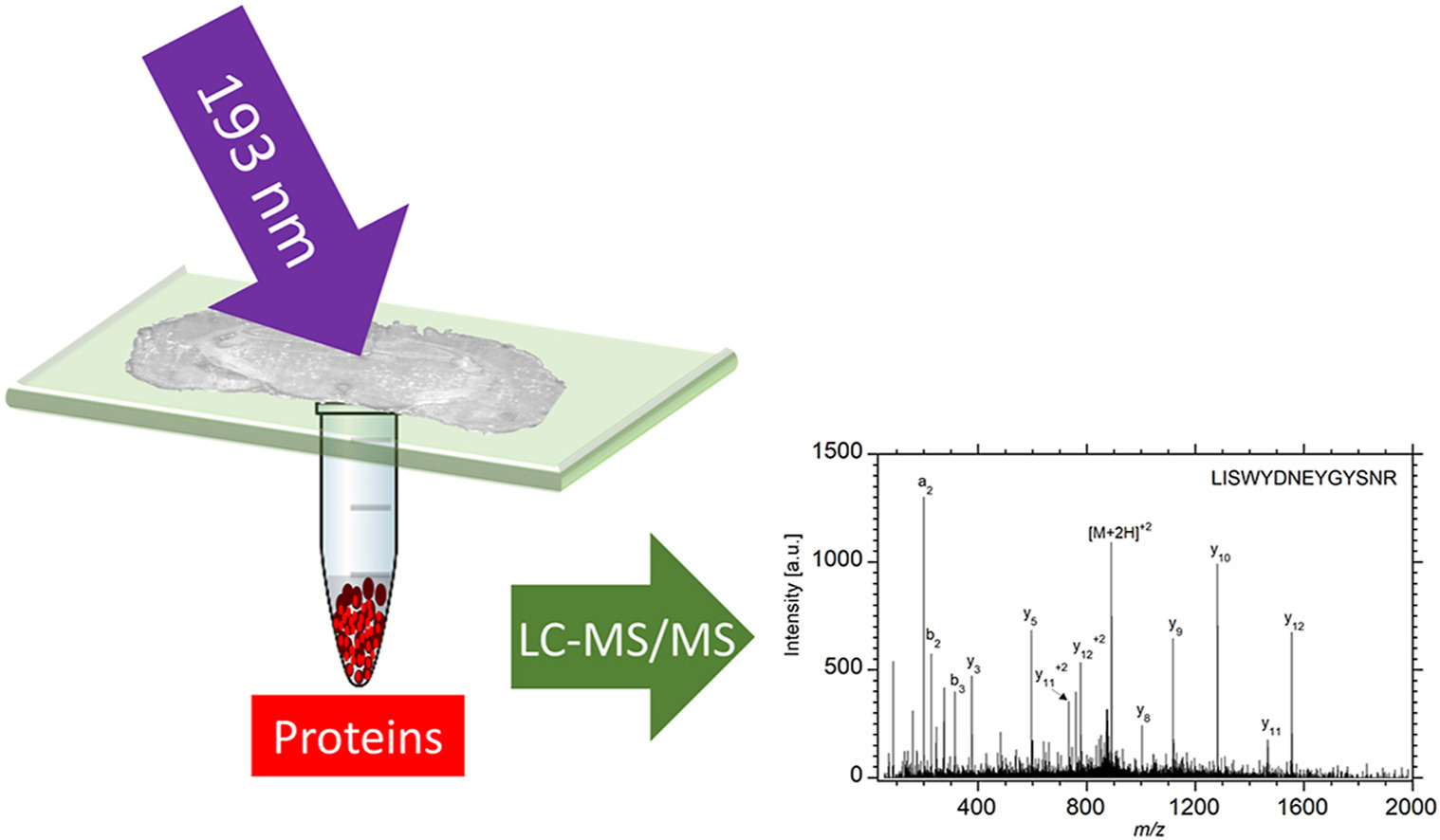
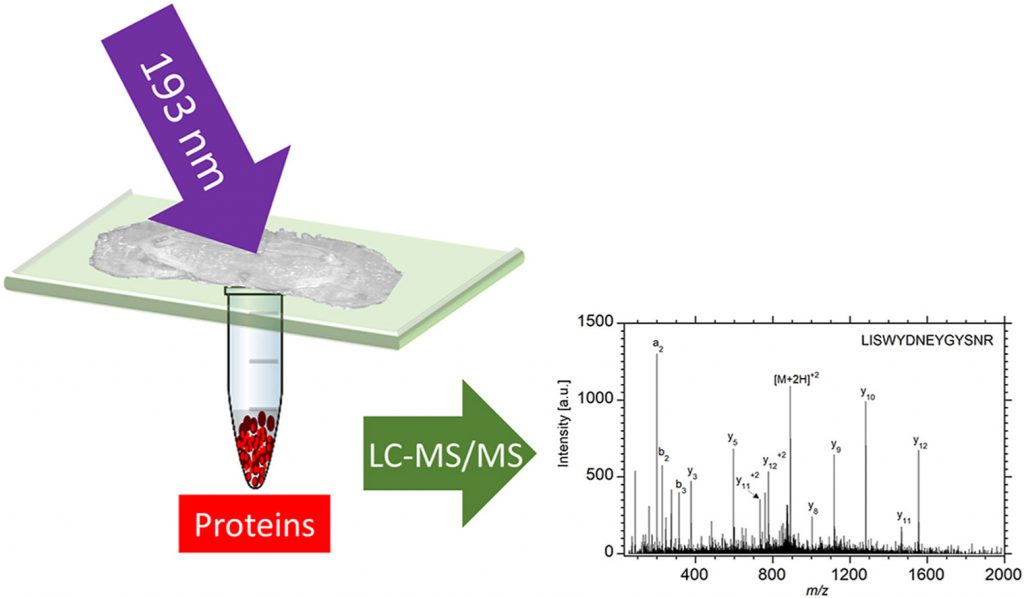
Deep-ultraviolet laser ablation with a pulsed 193 nm ArF excimer laser was used to remove localized regions from tissue sections from which proteins were extracted for spatially resolved proteomic analysis by liquid chromatography tandem mass spectrometry (LC-MS/MS). The ability to capture intact proteins by ablation at 193 nm wavelength was verified by matrix-assisted laser desorption ionization (MALDI) of the protein standard bovine serum albumin (BSA), which showed that BSA was ablated and captured without fragmentation. A Bradford assay of the ablated and captured proteins indicated 90% efficiency for transfer of the intact protein at a laser fluence of 3 kJ/m2. Rat brain tissue sections mounted on quartz microscope slides and ablated in transmission mode yielded 2 μg protein per mm2 as quantified by the Bradford assay. Tissue areas ranging from 0.06 mm2 to 1 mm2 were ablated and the ejected material was collected for proteomic analysis. Extracted proteins were digested and the resulting peptides were analyzed by LC-MS/MS. The proteins extracted from the ablated areas were identified and the average number of identified proteins ranged from 85 in the 0.06 mm2 area to 2400 in the 1 mm2 area of a 50 μm thick tissue. In comparison to infrared laser ablation of equivalent sampled areas, both the protein mass and number of proteins identified using DUV laser ablation sampling were approximately four times larger.