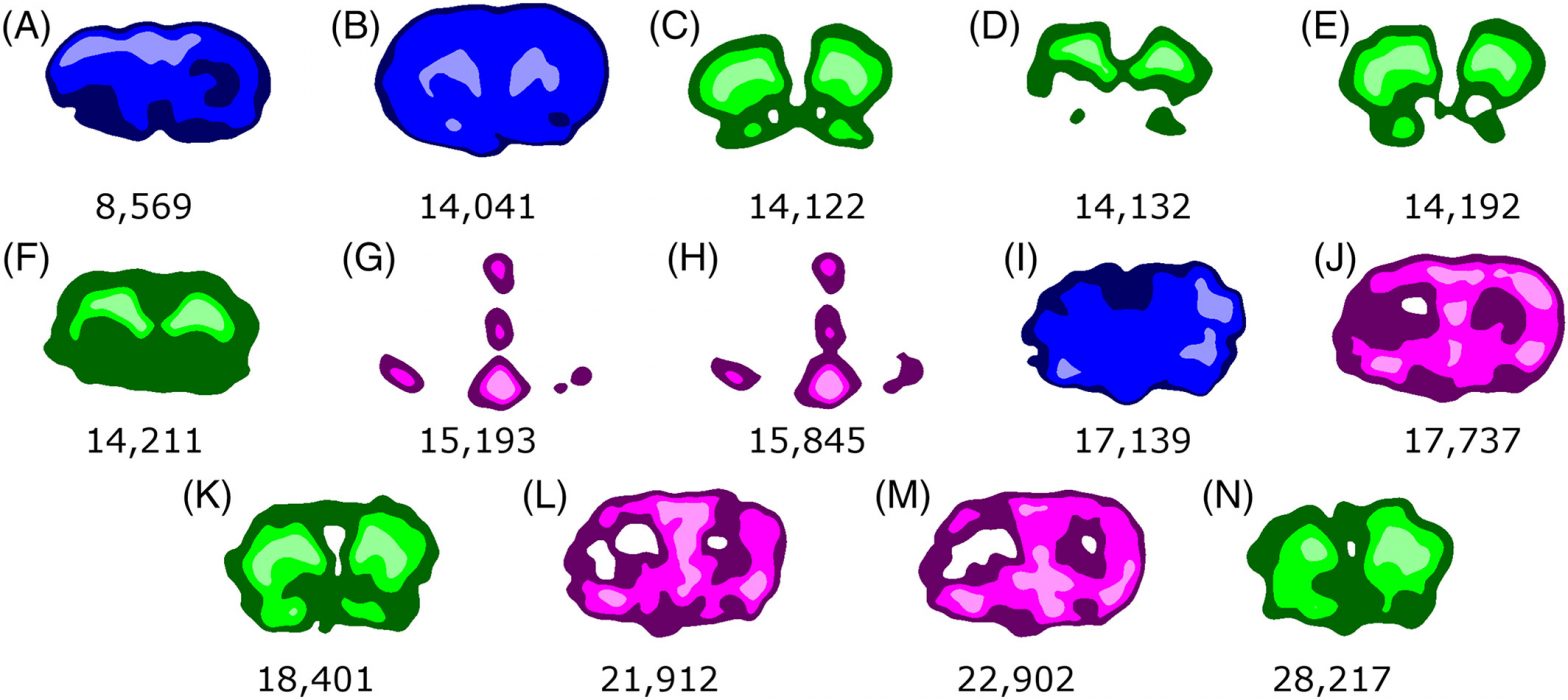
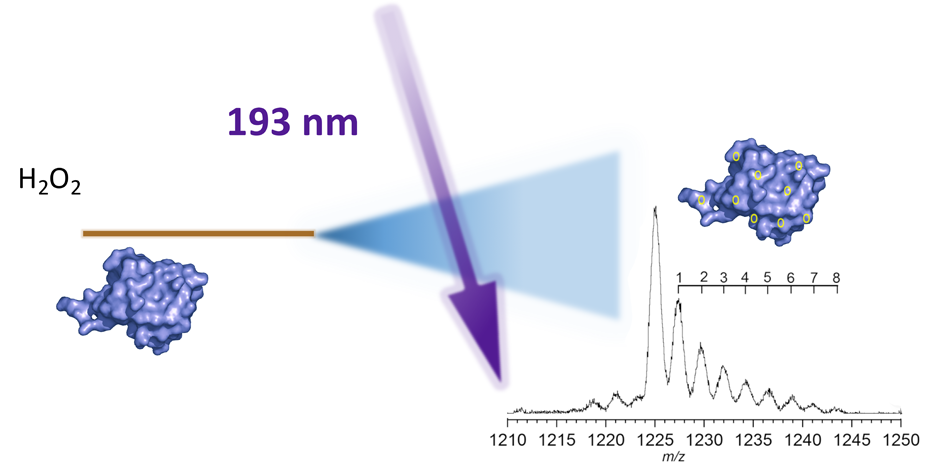
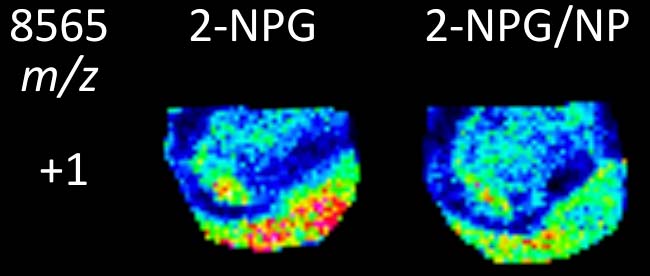
A.C. Pulukkody, Y.P. Yung, F. Donnarumma, K.K. Murray, R.P. Carlson, L. Hanley, Spatially resolved analysis of Pseudomonas aeruginosa biofilm proteomes measured by laser ablation sample transfer, PLoS One, 16 (2021) e0250911, doi: 10.1371/journal.pone.0250911 Abstract Heterogeneity in the distribution of nutrients and oxygen gradients during biofilm growth gives rise to changes in phenotype. There has been …
Category Archives: Publication
Lasers for matrix-assisted laser desorption ionization
K.K. Murray, Lasers for matrix-assisted laser desorption ionization, J. Mass Spectrom., 56 (2021) e4664. https://doi.org/10.1002/jms.4664 Abstract Matrix-assisted laser desorption ionization (MALDI) was introduced 35 years ago and has advanced from a general method for producing intact ions from large biomolecules to wide use in applications ranging from bacteria identification to tissue imaging. MALDI was enabled …
Continue reading “Lasers for matrix-assisted laser desorption ionization”
In defense of the quasimolecular ion
K.K. Murray, In defense of the quasimolecular ion, J. Mass Spectrom., 56 (2021) e4700; doi: 10.1002/jms.4700. Abstract The term quasimolecular ion has been used to describe ions comprising a molecule and weakly bound positive or negative ion or an ion formed by the loss of a proton from a molecule. This term was used in …
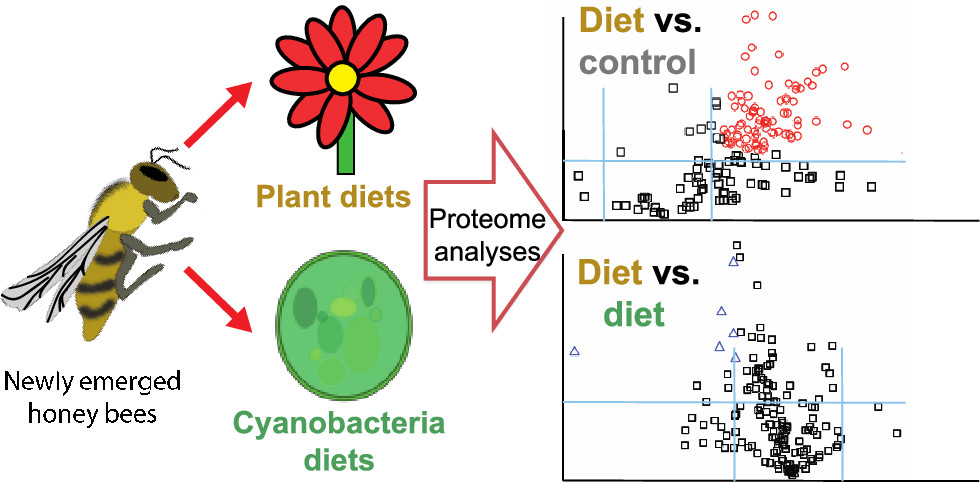
Honey Bee Proteome Responses to Plant and Cyanobacteria (blue-green algae) Diets
Abstract
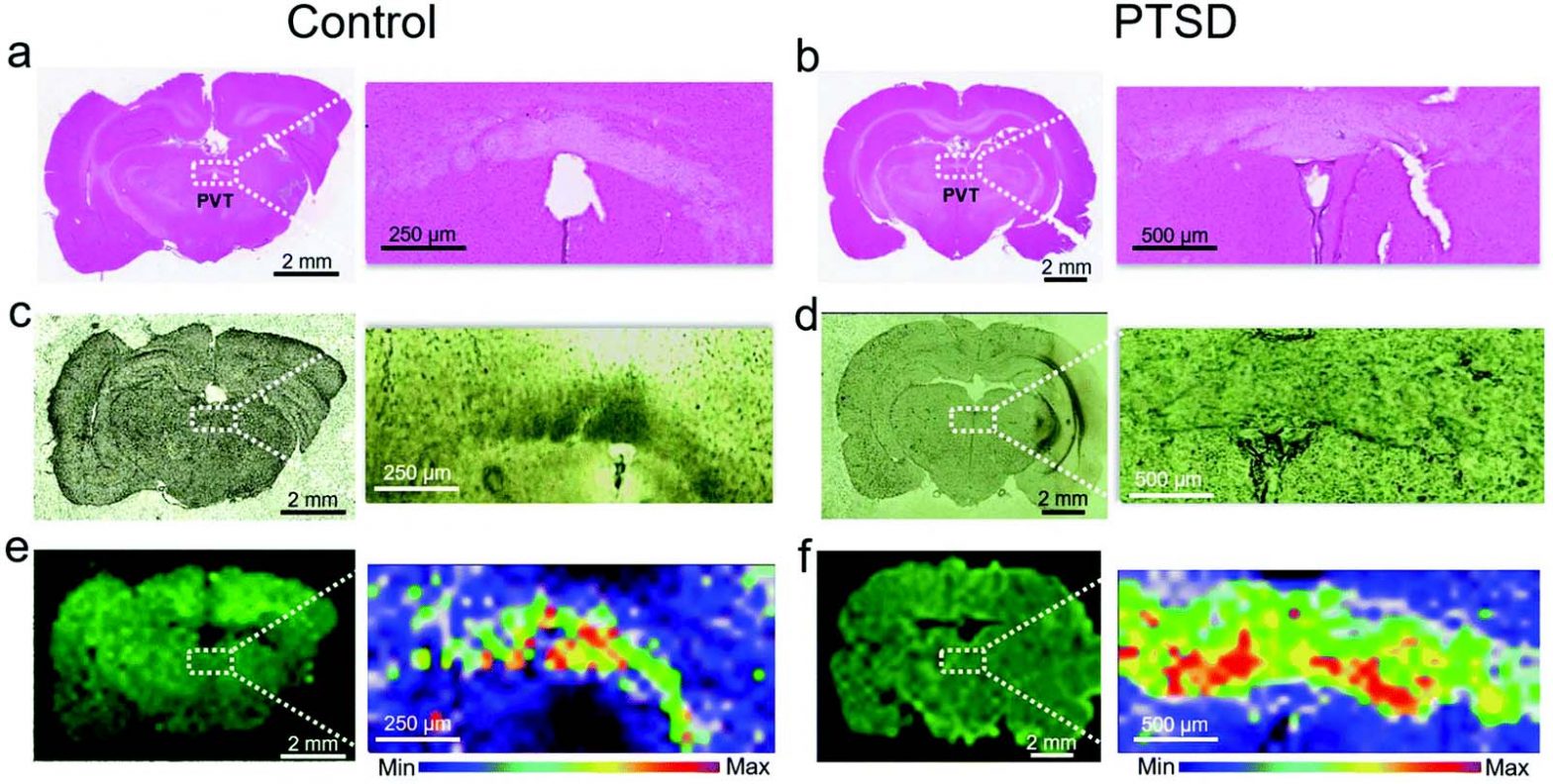
Label-free lipidome study of PVT of rat brain with post-traumatic stress injury by Raman imaging
A. Chaichi, S.M.A. Hasan, N. Mehta, F. Donnarumma, P. Ebenezer, K.K. Murray, J. Francis, M.R. Gartia, Label-free lipidome study of paraventricular thalamic nucleus (PVT) of rat brain with post-traumatic stress injury by Raman imaging, Analyst, 146 (2021) 170-183. Abstract Post-traumatic stress disorder (PTSD) is a widespread psychiatric injury that develops serious life-threatening symptoms like substance …
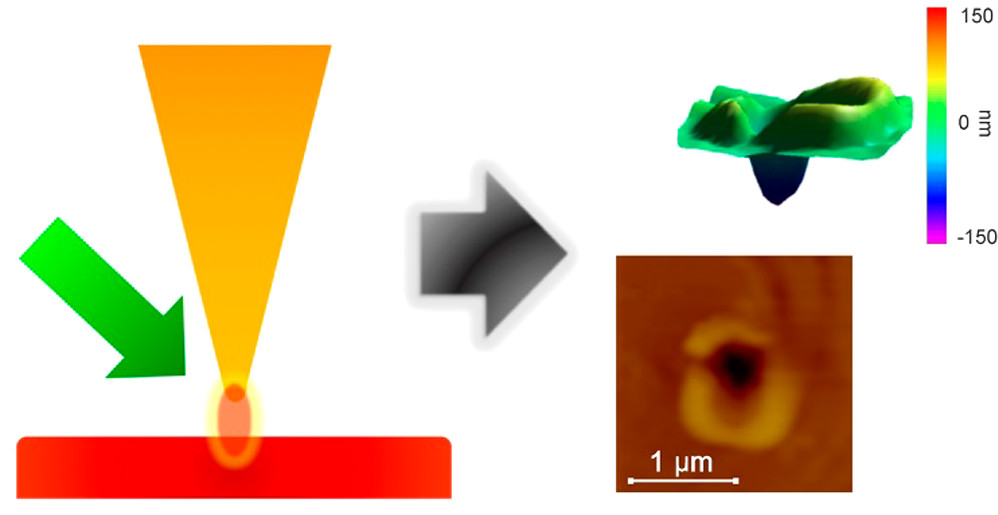
Wavelength-Dependent Tip-Enhanced Laser Ablation of Organic Dyes
F. Cao, F. Donnarumma, K.K. Murray, Wavelength-Dependent Tip-Enhanced Laser Ablation of Organic Dyes, Journal of Physical Chemistry C, 124 (2020) 1918-1922; doi: 10.1021/acs.jpcc.9b08081 Abstract The wavelength dependence of atomic force microscope apertureless tip-enhanced laser ablation was studied using a series of organic dyes to assess the effect of surface optical absorption. An optical parametric oscillator …
Continue reading “Wavelength-Dependent Tip-Enhanced Laser Ablation of Organic Dyes”
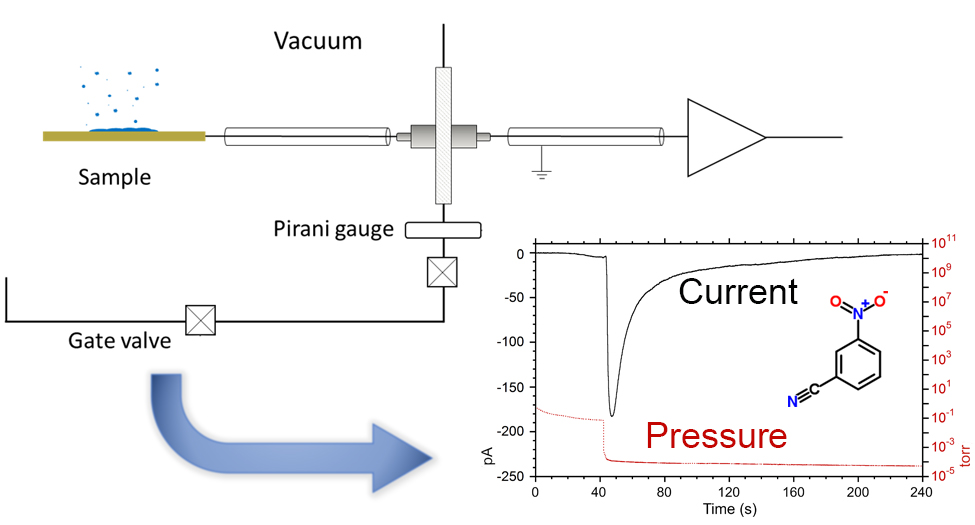
Sublimation Electrification of Organic Compounds
B. Banstola, K.K. Murray, Sublimation Electrification of Organic Compounds, J. Am. Soc. Mass. Spectrom., 31 (2020) 888-893; doi: 10.1021/jasms.9b00124. Abstract The electrification of crystalline deposits of organic compounds under high vacuum was measured and quantified. A group of compounds that produce multiply charged ions by matrix-assisted ionization were deposited on a metal plate, and the …
Continue reading “Sublimation Electrification of Organic Compounds”
MALDI imaging directed laser ablation tissue microsampling for data independent acquisition proteomics
K. Wang, F. Donnarumma, M.E. Pettit, C.W. Szot, T. Solouki, K.K. Murray, MALDI imaging directed laser ablation tissue microsampling for data independent acquisition proteomics, J. Mass Spectrom., 55 (2020) e4475; doi: 10.1002/jms.4475 Abstract A multimodal workflow for mass spectrometry imaging was developed that combines MALDI imaging with protein identification and quantification by liquid chromatography tandem …