H. Xia, K. Murray, S. Soper, J. Feng, Ultra sensitive affinity chromatography on avidin-functionalized PMMA microchip for low abundant post-translational modified protein enrichment, Biomed. Microdevices, 14 (2012) 67-81; doi: 10.1007/s10544-011-9586-7
Abstract
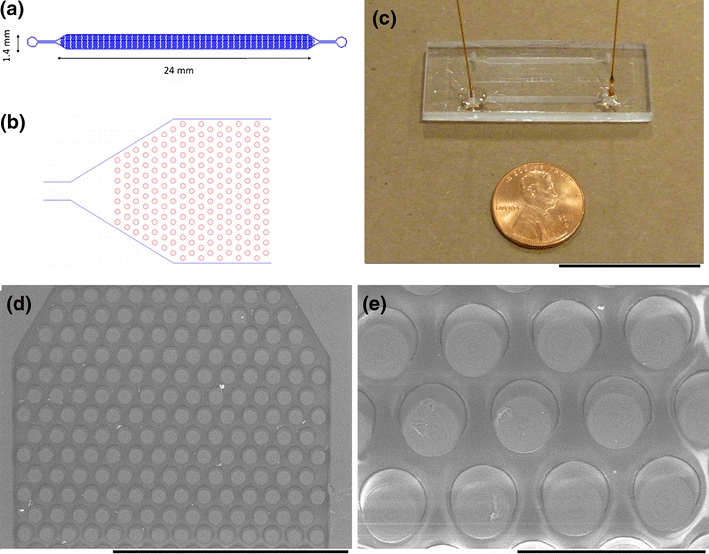
Post-translational modifications (PTM) of proteins play essential roles in cellular physiology and disease. The identification of protein substrates and detection of modification site helps understand PTM-mediated regulation in essential biological pathways and functions in various diseases. However, PTM proteins are typically present only at trace levels, making them difficult to identify in mass spectrometry based proteomics. In this paper, we report a novel and sensitive affinity chromatography on the avidin-functionalized poly(methyl methacrylate) (PMMA) microchip for enrichment of nanogram (ng) amount of PTMs. The chemical modification of poly(methyl methacrylate) (PMMA) surfaces yield avidin-terminated PMMA surfaces after UV radiation and consecutive EDC mediated coupling (amide reaction). This functionalized PMMA micro-device was developed to identify and specifically trap biotinylated PTM proteins of low abundance from complex protein mixture. Here we selected carbonylated protein as a representative PTM to illustrate the wide application of this affinity microchip for any PTMs converted into a tractable tag after derivatization. The surface topography, surface functional group mapping and elemental composition changes after each modification step of the treatment process were systematically measured qualitatively and quantitatively by atomic force microscopy, X-ray photoelectron spectroscopy and fluorescence microscopy. Quantitative study of biotinlated carbonylated protein capture recovery and elution efficiency of the device was also studied. We also envision that this subproteome enrichment micro-device can be assembled with other lab-on-a-chip components for follow-up protein analysis.