Link Disclosed are various embodiments for transferring molecules from a surface for mass spectrometry and other sample analysis methods, and the like. A laser is focused onto a tip of an atomic force microscope to remove and capture a quantity of molecules from the surface, so they can be transferred to a mass spectrometer or …
Tag Archives: Publication
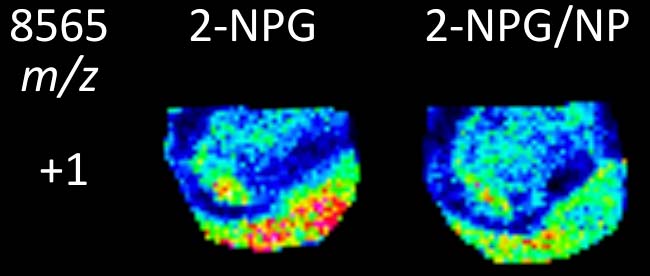
A Nanoparticle Co-matrix for Multiple Charging in MALDI Imaging of Tissue
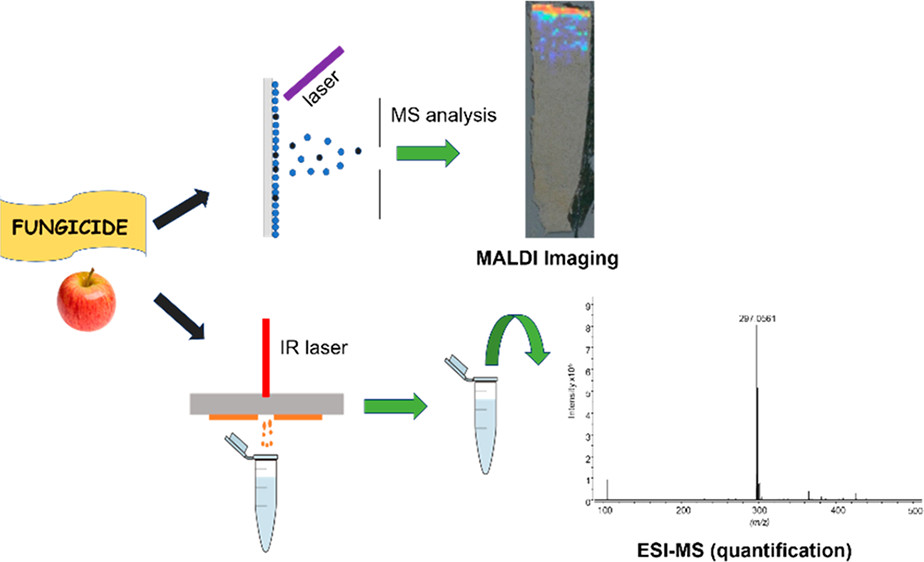
Matrix-Assisted Laser Desorption Ionization Imaging and Laser Ablation Sampling for Analysis of Fungicide Distribution in Apples
Piezoelectric matrix-assisted ionization
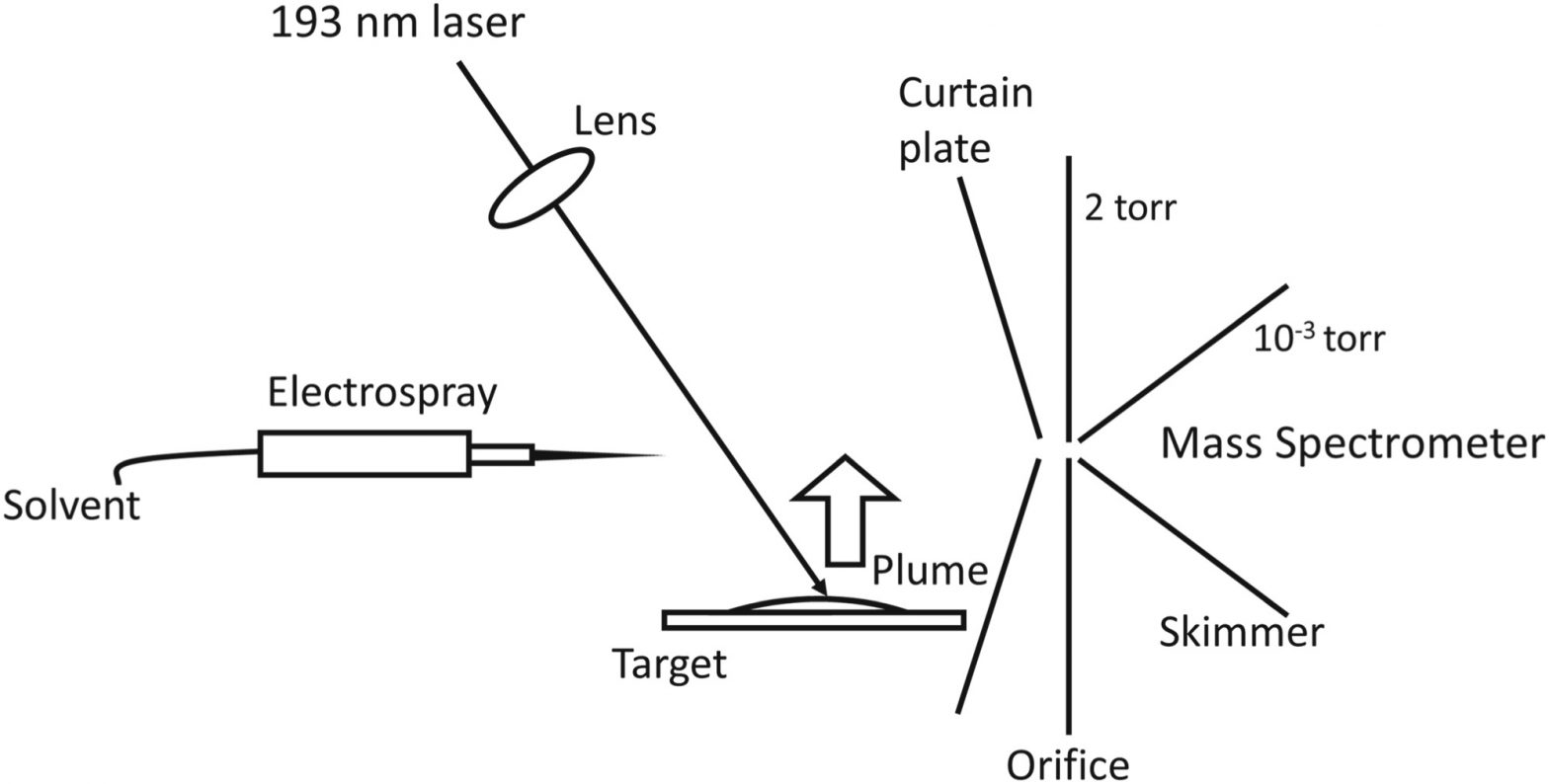
Deep‐ultraviolet laser ablation electrospray ionization mass spectrometry
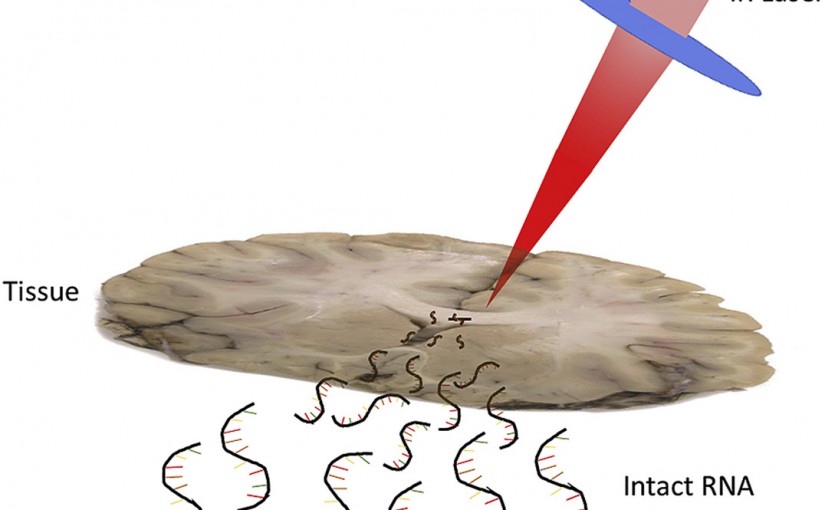
RNA Sampling from Tissue Sections using Infrared Laser Ablation
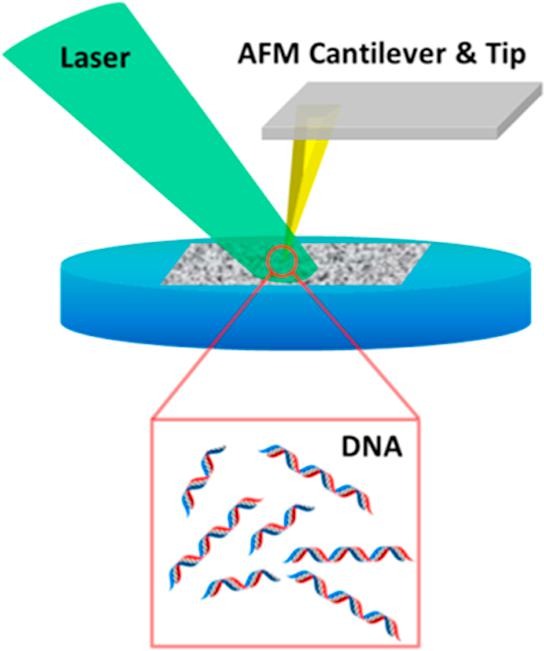
Tip-enhanced laser ablation and capture of DNA
Broadband ion mobility deconvolution for rapid analysis of complex mixtures
M.E. Pettit, M.R. Brantley, F. Donnarumma, K.K. Murray, T. Solouki, Broadband ion mobility deconvolution for rapid analysis of complex mixtures, Analyst. 143 (2018) 2574–2586. doi:10.1039/c8an00193f.
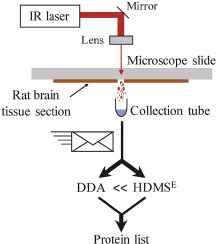
Infrared laser ablation sampling coupled with data independent high resolution UPLC-IM-MS/MS for tissue analysis
Infrared laser ablation and capture of enzymes with conserved activity
Wang, K., Donnarumma, F., Baldone, M. D., & Murray, K. K. Infrared laser ablation and capture of enzymes with conserved activity. Anal Chim Acta, 1027, 41–46 (2018). Abstract Infrared (IR) laser ablation at 3 μm wavelength was used to extract enzymes from tissue and quantitatively determine their activity. Experiments were conducted with trypsin, which was ablated, …
Continue reading “Infrared laser ablation and capture of enzymes with conserved activity”