J. Lee, S.A. Soper, K.K. Murray, “Development of an efficient on-chip digestion system for protein analysis using MALDI-TOF MS,” Analyst. 134 (2009) 2426–2433. doi:10.1039/b916556h.
Abstract

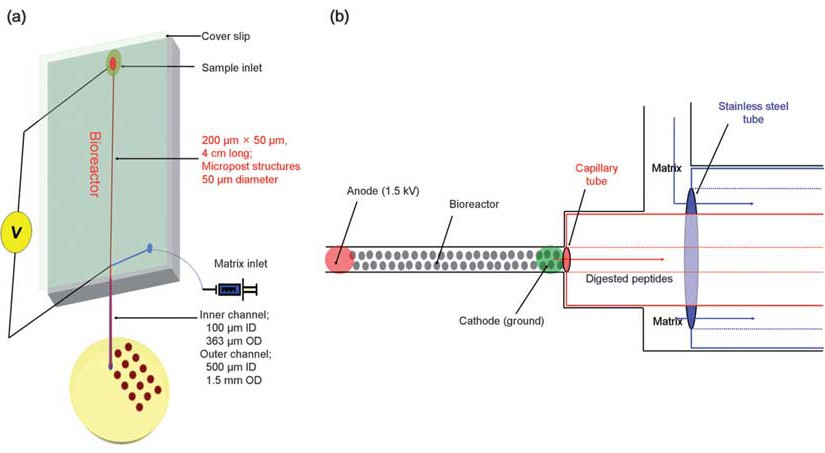
A solid-phase trypsin microreactor was constructed and operated with electrokinetically-driven flow for the digestion of proteins and coupled off-line with MALDI-TOF MS. The bioreactor was fabricated from poly(methyl methacrylate), PMMA, by hot embossing using a mold master prepared by micro-milling. The solid-phase bioreactor consisted of a 4 cm long, 200 microm wide, and 50 µm deep microfluidic channel that was populated with an array of 50 µm diameter micropost structures with a 50 µm inter-post spacing. The bioreactor was prepared by covalently attaching the proteolytic enzyme, trypsin, to the UV-modified surface of the PMMA microstructures using the appropriate coupling reagents. The performance of the system was evaluated using a set of proteins. The bioreactor provided efficient digestion of cytochrome c at a field strength of 375 V/cm, producing a reaction time of approximately 20 s to produce 97% sequence coverage for protein identification. Bovine serum albumin (BSA), phosphorylase b, and beta-casein were also assessed and the sequence coverages were 46, 63, and 79%, respectively, using the same reactor residence time. Furthermore, Escherichia coli was used as a model to demonstrate the feasibility of fingerprint analysis for intact cells using this solid-phase bioreactor.